Research progress of antibiotic resistance genes in wastewater treatment plants
-
摘要:
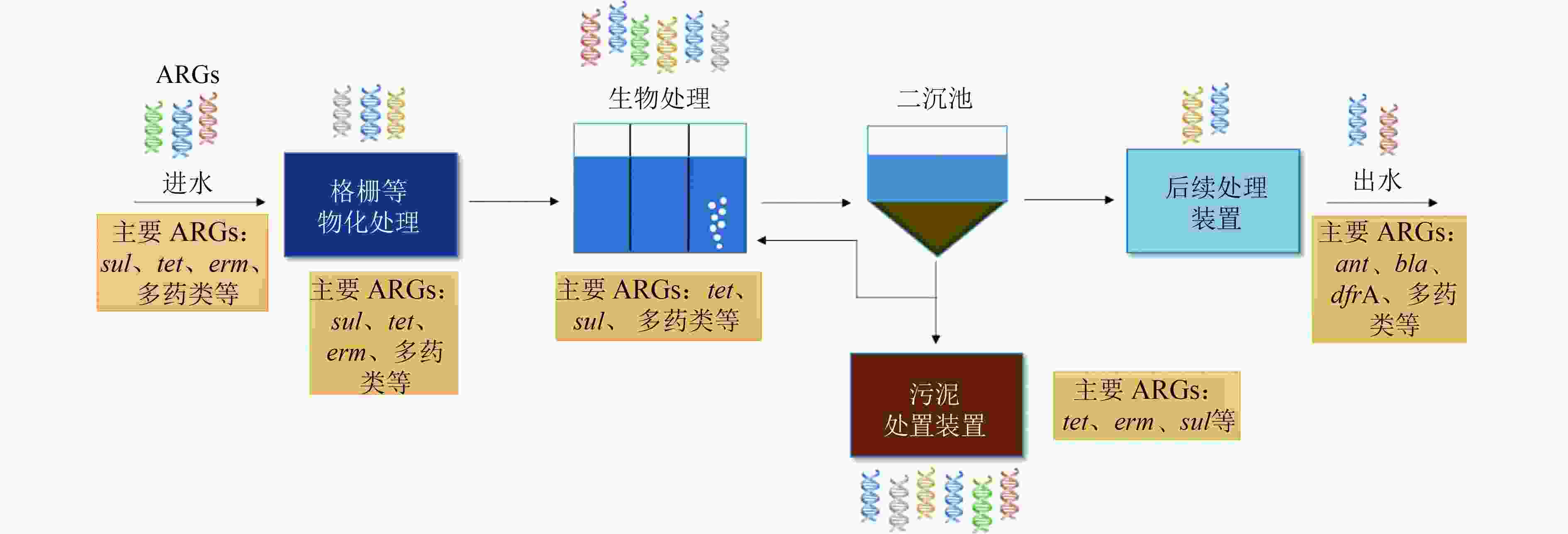
抗生素抗性基因(ARGs)是一类对自然环境及人体健康造成极大威胁的新型污染物,城市污水处理厂是ARGs的重要源和汇,具有重大潜在生态风险。系统梳理了污水处理过程中不同类型ARGs的组成变化特征和转移机制,提出β-内酰胺类、大环内酯类、四环素类、磺胺类、氨基糖苷类等类型ARGs广泛存在于全球污水处理厂中,但不同类型ARGs的丰度随污水处理过程的变化特征各异,且不同处理单元中的高丰度ARGs存在差异,水平转移是ARGs的主要转移机制。总结了环境条件、进水水质、操作参数等常见因素对ARGs丰度和分布的影响。在此基础上提出,识别具有指示作用的ARGs及其关键影响因素,定量分析各类因素对ARGs丰度、种类及水平转移机制的影响,以及建立ARGs风险评价标准体系是城市污水处理厂监测与控制ARGs潜在生态风险的未来发展方向。
Abstract:Antibiotic resistance genes (ARGs) are emerging contaminants which pose a great threat to the environment and human health. As an important source and sink of ARGs, wastewater treatment plants (WWTPs) have great potential ecological risks. Therefore, the diversity, composition as well as transfer mechanism of ARGs in the wastewater treatment processes were systematically introduced. ARGs of beta-lactam, macrolide, tetracycline, sulfonamide and aminoglycoside were widely detected in global WWTPs. However, the abundance of different types of ARGs along treatment processes and dominant ARGs detected in each process were different. Horizontal transfer was the main transfer mechanism of ARGs. The effects of common factors such as environmental conditions, influent wastewater quality and operational parameters on the abundance and distribution of ARGs were also summarized. It was proposed that the focus of future research was identifying the representative ARGs and the most influencing factors, quantifying the effects of various factors on the abundance, compositions and horizontal transfer mechanisms of ARGs, and establishing a standard system for evaluating the risks of ARGs for monitoring and controlling the potential ecological risks of ARGs in WWTPs.
-
表 1 不同地区污水处理厂中不同种类ARGs在进水、出水、污泥中的丰度变化
Table 1. Variations of abundance of different kinds of ARGs in influent, effluent and sludge in various WWTPs
抗性
基因
种类所在地区 抗性基因
亚型名称进水中丰度/
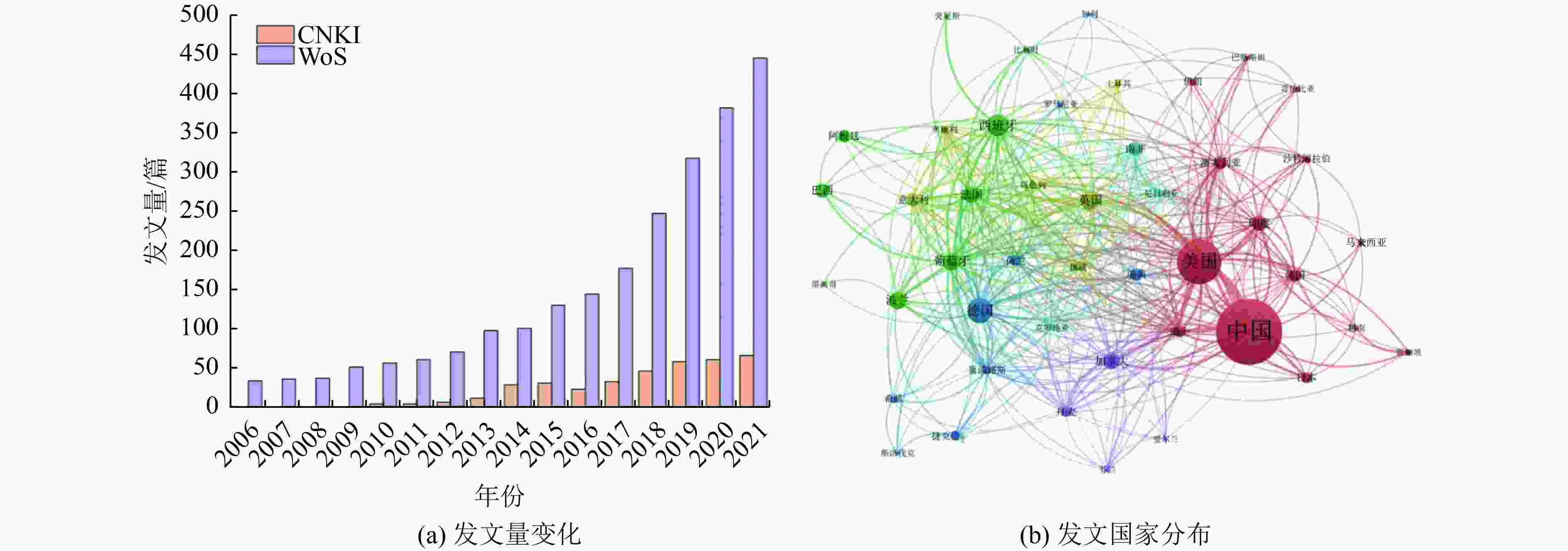
(拷贝/L)出水中丰度/
(拷贝/L)污泥中丰度/
(拷贝/g)去除
效果
(lg C)1)处理工艺 β-内酰胺类 中国北京[20] bl2d_oxa10、bl3_imp等51种 1.25×1010 4.73×107 1.47×1010 2.42 MBR工艺 美国加利福尼
亚州[21]blaM-1 2.23×108 2.82×106 1.9 活性污泥法+氯消毒 中国河北[22] blaPSE-1 106 2.46×104 107 2.64 A2/O工艺+氯消毒 美国马萨
诸塞州[21]blaTEM-uni 1.41×109 4.73×108 0.47 活性污泥法+氯消毒 瑞典哥德堡[20] mecA 5.01×104 5.01×103 1 活性污泥法+生物滤池 大环内酯类 中国北京[20] ermF、Inua等46种 1.17×1010 1.89×107 0.75×1010 2.79 MBR工艺 中国华北[23] erm 2种 (7.0±12)×107 (1.2±0.9)×1010 活性污泥法+氯消毒 中国河北[23] ermB 3×108 5.30×105 4×108 2.77 A2/O工艺+氯消毒 以色列夏夫丹[21] ermB 3.02×1010 2×105~3.02×107 3~4 活性污泥法 ermF 6.02×1010 3.02×106~2×108 2~4 四环素类 中国北京[20] tetG、tetM等
39 种1.39×1010 5.14×107 2.5×1010 2.43 MBR工艺 中国华北[22] tet 15种 (8.4±2.4)×107 (1.3±1.6)×1010 活性污泥法+氯消毒 中国香港[21] tetA 1010.78~1011.2 ND~107.33 3~4 活性污泥法+氯消毒 tetC 1011.13~1011.3 ND~107.12 4~5 中国南京[21] tetA 5.01×1010 1.41×109 1.55 活性污泥法 tetC 8.13×1010 1.38×109 1.77 中国河北[23] tetC 8×108 4. 13×106 2.23 A2/O工艺+氯消毒 美国威斯
康星州[21]tetG 109.4~1010.8 107.2~108.9 1~4 活性污泥法+紫外/氯消毒 tetQ 1010.2~1012 106.9~109.2 1~6 中国合肥[24] tetQ 2.03×1011 2×107 1.11×1010 4.01 SBR工艺+氯消毒 以色列夏夫丹[21] tetO 2×1010 ND~106 4.3 活性污泥法 美国密歇根州[25] tetO 5.13×109 9.12×106 1.78×109 2.75 活性污泥法+氯消毒 tetW 5.13×109 5.13×106 5.62×108 3 中国合肥[24] tetO 2×109 106 2×109 3.3 SBR+氯消毒 tetW 2×109 2×106 2×109 3 磺胺类 中国北京[20] dfrA1等7种 2.8×109 4.3×107 2.5×1010 1.81 MBR 中国浙江[26] dfrA1 1.3×107 2×105 9.38 ×105 1.9 A2/O dfrA13 8×106 1.2×105 5.93×104 1.8 中国华北[22] sul 3种 (6.7±7.2)×108 (2.2±2.8)×1011 活性污泥法+氯消毒 美国密歇根州[25] sul1 1.82×109 1.05×107 1.00×108 2.24 活性污泥法+氯消毒 美国密歇根州[21] sul1 108.46~1010.54 107.37~109.75 1~3 活性污泥法/氧化沟/生物转盘/MBR+紫外/氯消毒 中国合肥[24] sul1 3.88×1010 1.5×107 9.06×1010 3.41 SBR+氯消毒 sul2 3×109 4×106 109 2.88 以色列夏夫丹[21] sul1 1011 107.78~108.48 3~4 活性污泥法 sul2 1011 106.48~107.88 3~5 中国河北[22] sul2 2×108 5. 58×106 1010 1.7 A2/O+氯消毒 氨基糖苷类 中国北京[20] ant2ia、ant3ia等35种 3.26×1010 2.06×108 4.38×1010 2.2 MBR 氟喹诺酮-喹诺酮-氟苯尼考-氯霉素和安非霉素类 中国北京[20] cml_e3、catb3等9种 3.26×1010 0.84×107 1.67×1010 3.59 MBR 中国浙江[26] floR 1.2×107 2.1×105 1.59 × 105 1.8 A2/O 中国华北[22] qnr 3种 (7.3±9.6)×106 (1.5±2.3)×109 活性污泥法+氯消毒 多药类 中国北京[20] qacEdelta1、qacH等51种 1.39×1010 1.5×108 3.75×1010 1.97 MBR 中国哈尔滨[27] mexF 7.09×107 3×106 1.37 A/O 万古霉素类 比利时托里勒[21] vanA ND ND 活性污泥法+膜滤 中国哈尔滨[27] vanCO3 3.50×105 ND 5.54 A/O vanXD 1.08×104 4×103 0.43 1)C为丰度。注:ND表示未检出。 表 2 污水处理厂中ARGs分布和转移的主要影响因素
Table 2. Main influencing factors of ARGs distribution and transfer in WWTPs
影响因素 对ARGs的影响 文献来源 抗生素 磺胺类药物与sul1、四环素与tetX具有强相关性 [47-48] 选择作用具有交叉性,氯霉素与氨基糖苷类ARGs共现 [8,49] 氨苄青霉素造成大肠杆菌内多种非β-内酰胺类ARGs共现 [50] 高浓度的环丙沙星未造成特定耐药基因的富集,或存在滞后性与共选择 [51-52] 杀菌抗生素对ARGs的富集效果比抑菌抗生素更强 [53] 不同浓度的四环素影响ARGs的HGT [54] 重金属 水中抗生素抗性检出频率随重金属暴露浓度的升高而升高 [55] ARGs的丰度与钒等重金属浓度的相关性强于抗生素,湖泊重金属污染增强了细菌的耐药性 [60] 部分HMRGs与intI1基因具有相关性,影响ARGs的HGT [62] 其他有机物 甲苯、乙苯、PNP、PAP等芳香族化合物浓度影响ARGs [63] 苯乙烯、孔雀绿染料等能提升RP4质粒的转移效率 [60] 卡马西平等有机物通过增加活性氧(ROS)等机制促进ARGs的HGT [57] 纳米颗粒可能促进ARGs的富集传播,抑或去除ARGs [64-65] 微塑料促进污水处理厂中ARGs的富集与传播 [66-67] 微塑料对eARGs的吸附作用及促进HGT作用强于iARGs [68] 微塑料对二沉池出水ARGs的富集能力显著高于进水与污泥 [69] 环境因素及
水质条件温度与污水处理厂ARGs丰度显著相关,其中与bla、mcr丰度成反比,与ermB、sul2丰度成正比 [70-73] 温度影响ARGs的转移,夏季有利于VGT,冬季有利于HGT [28] 氨氮浓度与tetC、ermB等ARGs丰度呈正相关 [23] 高COD促进耐药菌繁殖,提高转化几率 [74] 盐度增加到1%以上会大大降低ARGs的总体丰度 [75] 操作参数及
处理工艺ARGs丰度与HRT呈正相关,与MLSS浓度、DO浓度、SRT呈负相关 [18] 高MLSS浓度、长SRT、低污泥负荷可降低ARGs丰度和多样性 [26,66-67,76] 厌氧与好氧工艺对ARGs影响较大 [77] 膜滤、电化学消毒、微生物燃料电池能显著降低ARGs [53,78-79] 氯消毒等后续处理对ARGs去除效果不一 [25,81-83] 氯及UV消毒会促进宿主繁殖,导致eARGs增多 [80] UV与氯消毒联用比单独氯消毒工艺效果好 [84] UV-AOPs自由基能够极大去除ARGs [86] AOPs中Fenton有关技术对ARGs去除效果优于生物处理及其他AOPs [85,87] 需调节改进工艺以保证ARB与ARGs的去除效果 [88-89] -
[1] 陈宇, 许亚南, 庞燕.抗生素赋存、来源及风险评估研究进展[J]. 环境工程技术学报,2021,11(3):562-570. doi: 10.12153/j.issn.1674-991X.20200180CHEN Y, XU Y N, PANG Y. Advances in research on the occurrence, source and risk assessment of antibiotics[J]. Journal of Environmental Engineering Technology,2021,11(3):562-570. doi: 10.12153/j.issn.1674-991X.20200180 [2] 申思奇. 交互带抗生素抗性基因污染分布特征及演变过程模拟研究[D]. 西安: 长安大学, 2021. [3] TAN L, WANG F, LIANG M M, et al. Antibiotic resistance genes attenuated with salt accumulation in saline soil[J]. Journal of Hazardous Materials,2019,374:35-42. doi: 10.1016/j.jhazmat.2019.04.020 [4] QIN K N, WEI L L, LI J J, et al. A review of ARGs in WWTPs: sources, stressors and elimination[J]. Chinese Chemical Letters,2020,31(10):2603-2613. doi: 10.1016/j.cclet.2020.04.057 [5] 宋冉冉, 国晓春, 卢少勇, 等.东洞庭湖表层水体中抗生素及抗性基因的赋存特征与源分析[J]. 环境科学研究,2021,34(9):2143-2153. doi: 10.13198/j.issn.1001-6929.2021.04.27SONG R R, GUO X C, LU S Y, et al. Occurrence and source analysis of antibiotics and antibiotic resistance genes in surface water of East Dongting Lake Basin[J]. Research of Environmental Sciences,2021,34(9):2143-2153. doi: 10.13198/j.issn.1001-6929.2021.04.27 [6] ZHANG A N, HOU C J, NEGI M, et al. Online searching platform for the antibiotic resistome in bacterial tree of life and global habitats[J]. FEMS Microbiology Ecology,2020,96(7):fiaa107. doi: 10.1093/femsec/fiaa107 [7] 王敏妍, 李亚丽, 邹世春, 等.水环境中胞内外抗生素抗性基因分析的DNA提取方法及污染现状研究进展[J]. 分析测试学报,2021,40(6):869-875. doi: 10.3969/j.issn.1004-4957.2021.06.012WANG M Y, LI Y L, ZOU S C, et al. DNA extraction methods for intracellular and extracellular antibiotic resistance genes and their pollution status in aquatic environment[J]. Journal of Instrumental Analysis,2021,40(6):869-875. doi: 10.3969/j.issn.1004-4957.2021.06.012 [8] 耿嘉璐. 抗性基因和药物的多介质环境分布特征与生态风险评价[D]. 哈尔滨: 哈尔滨工业大学, 2020. [9] ZHANG D W, PENG Y, CHAN C L, et al. Metagenomic survey reveals more diverse and abundant antibiotic resistance genes in municipal wastewater than hospital wastewater[J]. Frontiers in Microbiology,2021,12:712843. doi: 10.3389/fmicb.2021.712843 [10] JU F, LI B, MA L P, et al. Antibiotic resistance genes and human bacterial pathogens: co-occurrence, removal, and enrichment in municipal sewage sludge digesters[J]. Water Research,2016,91:1-10. doi: 10.1016/j.watres.2015.11.071 [11] PRUDEN A,PEI R T,STORTEBOOM H,et al. Antibiotic resistance genes as emerging contaminants:studies in northern Colorado[J]. Environmental Science & Technology,2006,40(23):7445-7450. [12] 严岩, 尤本胜, 刘伟京, 等.基于文献计量学的近20年水环境中抗生素污染研究趋势及热点分析[J]. 环境工程技术学报,2023,13(3):1161-1167. doi: 10.12153/j.issn.1674-991X.20220343YAN Y, YOU B S, LIU W J, et al. Research trend and hot spot analysis of antibiotic pollution in water environment in recent 20 years based on bibliometrics[J]. Journal of Environmental Engineering Technology,2023,13(3):1161-1167. doi: 10.12153/j.issn.1674-991X.20220343 [13] 付垚. 抗生素压力下好氧颗粒污泥的培养及抗性基因归趋[D]. 济南: 山东大学, 2020. [14] PAZDA M, KUMIRSKA J, STEPNOWSKI P, et al. Antibiotic resistance genes identified in wastewater treatment plant systems:a review[J]. Science of the Total Environment,2019,697:134023. doi: 10.1016/j.scitotenv.2019.134023 [15] WANG J L, CHU L B, WOJNÁROVITS L, et al. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: an overview[J]. The Science of the Total Environment,2020,744:140997. doi: 10.1016/j.scitotenv.2020.140997 [16] AUGUET O, PIJUAN M T, BORREGO C M, et al. Sewers as potential reservoirs of antibiotic resistance[J]. Science of the Total Environment,2017,605/606:1047-1054. doi: 10.1016/j.scitotenv.2017.06.153 [17] FAN X Y, GAO J F, PAN K L, et al. Functional genera, potential pathogens and predicted antibiotic resistance genes in 16 full-scale wastewater treatment plants treating different types of wastewater[J]. Bioresource Technology,2018,268:97-106. doi: 10.1016/j.biortech.2018.07.118 [18] AN X L, SU J Q, LI B, et al. Tracking antibiotic resistome during wastewater treatment using high throughput quantitative PCR[J]. Environment International,2018,117:146-153. doi: 10.1016/j.envint.2018.05.011 [19] SU J Q, AN X L, LI B, et al. Correction to: metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China[J]. Microbiome,2018,6(1):127. doi: 10.1186/s40168-018-0504-6 [20] ZHENG W L, HUYAN J Q, TIAN Z, et al. Clinical class 1 integron-integrase gene:a promising indicator to monitor the abundance and elimination of antibiotic resistance genes in an urban wastewater treatment plant[J]. Environment International,2020,135:105372. doi: 10.1016/j.envint.2019.105372 [21] HONG P Y, AL-JASSIM N, ANSARI M I, et al. Environmental and public health implications of water reuse: antibiotics, antibiotic resistant bacteria, and antibiotic resistance genes[J]. Antibiotics,2013,2(3):367-399. doi: 10.3390/antibiotics2030367 [22] MAO D Q, YU S, RYSZ M, et al. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants[J]. Water Research,2015,85:458-466. doi: 10.1016/j.watres.2015.09.010 [23] 谢辉, 包樱钰, 李菲菲, 等.A2/O生活污水处理系统中抗生素抗性基因的分布及去除[J]. 环境工程,2019,37(12):80-89. doi: 10.13205/j.hjgc.201912015XIE H, BAO Y Y, LI F F, et al. Distribution and removal of antibiotic resistance genes in an A2/O domestic wastewater treatment plant[J]. Environmental Engineering,2019,37(12):80-89. doi: 10.13205/j.hjgc.201912015 [24] YUAN L, LI Z H, ZHANG M Q, et al. Mercury/silver resistance genes and their association with antibiotic resistance genes and microbial community in a municipal wastewater treatment plant[J]. Science of the Total Environment,2019,657:1014-1022. doi: 10.1016/j.scitotenv.2018.12.088 [25] GAO P, MUNIR M, XAGORARAKI I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant[J]. Science of the Total Environment,2012,421/422:173-183. doi: 10.1016/j.scitotenv.2012.01.061 [26] 姚鹏城, 陈嘉瑜, 张永明, 等.抗生素抗性基因在生活及工业混合废水处理系统中的分布和去除[J]. 生态毒理学报,2020,15(1):201-208. doi: 10.7524/AJE.1673-5897.20190307004YAO P C, CHEN J Y, ZHANG Y M, et al. Distribution and removal of antibiotic resistance genes in municipal and industrial mixed wastewater treatment systems[J]. Asian Journal of Ecotoxicology,2020,15(1):201-208. doi: 10.7524/AJE.1673-5897.20190307004 [27] SUN S J, GENG J L, MA L X, et al. Changes in antibiotic resistance genotypes and phenotypes after two typical sewage disposal processes[J]. Chemosphere,2022,291:132833. doi: 10.1016/j.chemosphere.2021.132833 [28] CHE Y, XIA Y, LIU L, et al. Mobile antibiotic resistome in wastewater treatment plants revealed by Nanopore metagenomic sequencing[J]. Microbiome,2019,7(1):44. doi: 10.1186/s40168-019-0663-0 [29] 袁立霞. 制药废水处理中菌群特征与抗性基因传播规律研究[D]. 石家庄: 河北科技大学, 2019. [30] 金亦豪, 刘子述, 胡宝兰.环境中胞内胞外抗性基因的分离检测、分布与传播研究进展[J]. 微生物学报,2022,62(4):1247-1259.JIN Y H, LIU Z S, HU B L. Isolation, detection, distribution, and transmission of intracellular and extracellular antibiotic resistance genes in the environment[J]. Acta Microbiologica Sinica,2022,62(4):1247-1259. [31] YU K F, LI P, HE Y L, et al. Unveiling dynamics of size-dependent antibiotic resistome associated with microbial communities in full-scale wastewater treatment plants[J]. Water Research,2020,187:116450. doi: 10.1016/j.watres.2020.116450 [32] WRIGHT G D. Antibiotic resistance in the environment: a link to the clinic[J]. Current Opinion in Microbiology,2010,13(5):589-594. doi: 10.1016/j.mib.2010.08.005 [33] SU J Q, WEI B, OUYANG W Y, et al. Antibiotic resistome and its association with bacterial communities during sewage sludge composting[J]. Environmental Science & Technology,2015,49(12):7356-7363. [34] ZHOU J Z, KANG S, SCHADT C W, et al. Spatial scaling of functional gene diversity across various microbial taxa[J]. Proceedings of the National Academy of Sciences of the United States of America,2008,105(22):7768-7773. doi: 10.1073/pnas.0709016105 [35] MARTÍNEZ J L. Antibiotics and antibiotic resistance genes in natural environments[J]. Science,2008,321(5887):365-367. doi: 10.1126/science.1159483 [36] WEI Z Y, FENG K, WANG Z J, et al. High-throughput single-cell technology reveals the contribution of horizontal gene transfer to typical antibiotic resistance gene dissemination in wastewater treatment plants[J]. Environmental Science & Technology,2021,55(17):11824-11834. [37] SMALLA K, JECHALKE S, TOP E M. Plasmid detection, characterization, and ecology[J]. Microbiology Spectrum,2015,3(1):PLAS-0038-2014. [38] CZEKALSKI N, BERTHOLD T, CAUCCI S, et al. Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland[J]. Frontiers in Microbiology,2012,3:106. [39] 武彩云, 李慧莉, 覃彩霞, 等.螺旋霉素废水处理过程中菌群结构、水质特征及抗性基因之间关系分析[J]. 环境科学,2021,42(9):4358-4365. doi: 10.13227/j.hjkx.202101086WU C Y, LI H L, QIN C X, et al. Mutual influence between microbial community, wastewater characteristics, and antibiotic resistance genes during spiramycin production wastewater treatment[J]. Environmental Science,2021,42(9):4358-4365. doi: 10.13227/j.hjkx.202101086 [40] SEITZ P, BLOKESCH M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria[J]. FEMS Microbiology Reviews,2013,37(3):336-363. doi: 10.1111/j.1574-6976.2012.00353.x [41] WANG Y, LU J, ENGELSTÄDTER J, et al. Non-antibiotic pharmaceuticals enhance the transmission of exogenous antibiotic resistance genes through bacterial transformation[J]. The ISME Journal,2020,14(8):2179-2196. doi: 10.1038/s41396-020-0679-2 [42] ZHANG S, WANG Y, LU J, et al. Chlorine disinfection facilitates natural transformation through ROS-mediated oxidative stress[J]. The ISME Journal,2021,15(10):2969-2985. doi: 10.1038/s41396-021-00980-4 [43] COLOMER-LLUCH M, IMAMOVIC L, JOFRE J, et al. Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry[J]. Antimicrobial Agents and Chemotherapy,2011,55(10):4908-4911. doi: 10.1128/AAC.00535-11 [44] CALERO-CÁCERES W, MELGAREJO A, COLOMER-LLUCH M, et al. Sludge as a potential important source of antibiotic resistance genes in both the bacterial and bacteriophage fractions[J]. Environmental Science & Technology,2014,48(13):7602-7611. [45] ZHAO J H, LI B, LV P, et al. Distribution of antibiotic resistance genes and their association with bacteria and viruses in decentralized sewage treatment facilities[J]. Frontiers of Environmental Science & Engineering,2022,16(3):35. [46] FOGG P C M. Identification and characterization of a direct activator of a gene transfer agent[J]. Nature Communications,2019,10:595. doi: 10.1038/s41467-019-08526-1 [47] ZHENG W L, WEN X H, ZHANG B, et al. Selective effect and elimination of antibiotics in membrane bioreactor of urban wastewater treatment plant[J]. Science of the Total Environment,2019,646:1293-1303. doi: 10.1016/j.scitotenv.2018.07.400 [48] 刘航. 典型抗生素与污水脱氮除磷工艺微生物相互作用机理研究[D]. 天津: 天津大学, 2017. [49] ZHAO R X, YU K, ZHANG J Y, et al. Deciphering the mobility and bacterial hosts of antibiotic resistance genes under antibiotic selection pressure by metagenomic assembly and binning approaches[J]. Water Research,2020,186:116318. doi: 10.1016/j.watres.2020.116318 [50] LI A D, MA L P, JIANG X T, et al. Cultivation-dependent and high-throughput sequencing approaches studying the co-occurrence of antibiotic resistance genes in municipal sewage system[J]. Applied Microbiology and Biotechnology,2017,101(22):8197-8207. doi: 10.1007/s00253-017-8573-1 [51] BENGTSSON-PALME J, HAMMARÉN R, PAL C, et al. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics[J]. Science of the Total Environment,2016,572:697-712. doi: 10.1016/j.scitotenv.2016.06.228 [52] GAO Y X, LI X, FAN X Y, et al. Wastewater treatment plants as reservoirs and sources for antibiotic resistance genes: a review on occurrence, transmission and removal[J]. Journal of Water Process Engineering,2022,46:102539. doi: 10.1016/j.jwpe.2021.102539 [53] SHETH R U, CABRAL V, CHEN S P, et al. Manipulating bacterial communities by in situ microbiome engineering[J]. Trends in Genetics,2016,32(4):189-200. doi: 10.1016/j.tig.2016.01.005 [54] JUTKINA J, RUTGERSSON C, FLACH C F, et al. An assay for determining minimal concentrations of antibiotics that drive horizontal transfer of resistance[J]. Science of the Total Environment,2016,548/549:131-138. doi: 10.1016/j.scitotenv.2016.01.044 [55] STEPANAUSKAS R, GLENN T C, JAGOE C H, et al. Coselection for microbial resistance to metals and antibiotics in freshwater microcosms[J]. Environmental Microbiology,2006,8(9):1510-1514. doi: 10.1111/j.1462-2920.2006.01091.x [56] JIAO Y N, CHEN H, GAO R X, et al. Organic compounds stimulate horizontal transfer of antibiotic resistance genes in mixed wastewater treatment systems[J]. Chemosphere,2017,184:53-61. doi: 10.1016/j.chemosphere.2017.05.149 [57] WANG Y, LU J, MAO L K, et al. Antiepileptic drug carbamazepine promotes horizontal transfer of plasmid-borne multi-antibiotic resistance genes within and across bacterial Genera[J]. The ISME Journal,2019,13(2):509-522. doi: 10.1038/s41396-018-0275-x [58] MA Y J, METCH J W, YANG Y, et al. Shift in antibiotic resistance gene profiles associated with nanosilver during wastewater treatment[J]. FEMS Microbiology Ecology,2016,92(3):fiw022. doi: 10.1093/femsec/fiw022 [59] SONG J X, RENSING C, HOLM P E, et al. Comparison of metals and tetracycline as selective agents for development of tetracycline resistant bacterial communities in agricultural soil[J]. Environmental Science & Technology,2017,51(5):3040-3047. [60] KOMIJANI M, SHAMABADI N S, SHAHIN K, et al. Heavy metal pollution promotes antibiotic resistance potential in the aquatic environment[J]. Environmental Pollution,2021,274:116569. doi: 10.1016/j.envpol.2021.116569 [61] GUPTA S K, SHIN H, HAN D, et al. Metagenomic analysis reveals the prevalence and persistence of antibiotic- and heavy metal-resistance genes in wastewater treatment plant[J]. Journal of Microbiology,2018,56(6):408-415. doi: 10.1007/s12275-018-8195-z [62] Di CESARE A, ECKERT E M, D'URSO S, et al. Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants[J]. Water Research,2016,94:208-214. doi: 10.1016/j.watres.2016.02.049 [63] MA X Y, ZHANG X W, XIA J T, et al. Phenolic compounds promote the horizontal transfer of antibiotic resistance genes in activated sludge[J]. The Science of the Total Environment,2021,800:149549. doi: 10.1016/j.scitotenv.2021.149549 [64] CUI H L, SMITH A L. Impact of engineered nanoparticles on the fate of antibiotic resistance genes in wastewater and receiving environments: a comprehensive review[J]. Environmental Research, 2022, 204: 112373. [65] EZEUKO A S, OJEMAYE M O, OKOH O O, et al. Potentials of metallic nanoparticles for the removal of antibiotic resistant bacteria and antibiotic resistance genes from wastewater: a critical review[J]. Journal of Water Process Engineering,2021,41:102041. doi: 10.1016/j.jwpe.2021.102041 [66] YANG Y Y, LIU G H, SONG W J, et al. Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes[J]. Environment International,2019,123:79-86. doi: 10.1016/j.envint.2018.11.061 [67] SYRANIDOU E, KALOGERAKIS N. Interactions of microplastics, antibiotics and antibiotic resistant genes within WWTPs[J]. Science of the Total Environment,2022,804:150141. doi: 10.1016/j.scitotenv.2021.150141 [68] CHENG Y, LU J R, FU S S, et al. Enhanced propagation of intracellular and extracellular antibiotic resistance genes in municipal wastewater by microplastics[J]. Environmental Pollution, 2022, 292(Pt A): 118284. [69] 吴文斌, 付树森, 毛步云, 等.微塑料对城市污水中胞内和胞外抗性基因的富集特征研究[J]. 环境科学研究,2021,34(6):1434-1440. doi: 10.13198/j.issn.1001-6929.2021.02.15WU W B, FU S S, MAO B Y, et al. Enrichment of intracellular and extracellular antibiotic resistance genes by microplastics in municipal wastewater[J]. Research of Environmental Sciences,2021,34(6):1434-1440. doi: 10.13198/j.issn.1001-6929.2021.02.15 [70] SUN C X, ZHANG B, NING D L, et al. Seasonal dynamics of the microbial community in two full-scale wastewater treatment plants: diversity, composition, phylogenetic group based assembly and co-occurrence pattern[J]. Water Research,2021,200:117295. doi: 10.1016/j.watres.2021.117295 [71] ZHANG B, SUN C X, XIA Y, et al. Profiles of antibiotic resistance genes and virulence genes and their temporal interactions in the membrane bioreactor and oxidation ditch[J]. Environment International,2019,131:104980. doi: 10.1016/j.envint.2019.104980 [72] SCHAGES L, WICHERN F, KALSCHEUER R, et al. Winter is coming:impact of temperature on the variation of beta-lactamase and mcr genes in a wastewater treatment plant[J]. Science of the Total Environment,2020,712:136499. doi: 10.1016/j.scitotenv.2020.136499 [73] RODRÍGUEZ E A, PINO N J, JIMÉNEZ J N. Climatological and epidemiological conditions are important factors related to the abundance of blaKPC and other antibiotic resistance genes (ARGs) in wastewater treatment plants and their effluents, in an endemic country[J]. Frontiers in Cellular and Infection Microbiology,2021,11:686472. doi: 10.3389/fcimb.2021.686472 [74] JIN M, LIU L, WANG D N, et al. Chlorine disinfection promotes the exchange of antibiotic resistance genes across bacterial Genera by natural transformation[J]. The ISME Journal,2020,14(7):1847-1856. doi: 10.1038/s41396-020-0656-9 [75] LIU M T, LI Q L, SUN H H, et al. Impact of salinity on antibiotic resistance genes in wastewater treatment bioreactors[J]. Chemical Engineering Journal,2018,338:557-563. doi: 10.1016/j.cej.2018.01.066 [76] MUNIR M, WONG K, XAGORARAKI I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan[J]. Water Research,2011,45(2):681-693. doi: 10.1016/j.watres.2010.08.033 [77] CHRISTGEN B, YANG Y, AHAMMAD S Z, et al. Metagenomics shows that low-energy anaerobic-aerobic treatment reactors reduce antibiotic resistance gene levels from domestic wastewater[J]. Environmental Science & Technology,2015,49(4):2577-2584. [78] ANTHONY E T, OJEMAYE M O, OKOH A I, et al. Potentials of low-cost methods for the removal of antibiotic-resistant bacteria and their genes in low budget communities: a review[J]. Journal of Water Process Engineering,2021,40:101919. doi: 10.1016/j.jwpe.2021.101919 [79] 张启伟, 孙丽华, 史鹏飞, 等.混凝沉淀-UF工艺去除二级出水中ARGs效能研究[J]. 环境科学研究,2019,32(4):718-724. doi: 10.13198/j.issn.1001-6929.2018.09.25ZHANG Q W, SUN L H, SHI P F, et al. Removal efficiency of ARGs in secondary effluent by coagulation-sedimentation-UF process[J]. Research of Environmental Sciences,2019,32(4):718-724. doi: 10.13198/j.issn.1001-6929.2018.09.25 [80] 付树森, 王艺, 王肖霖, 等.氯和紫外消毒过程中胞外抗性基因的产生特征[J]. 中国环境科学,2021,41(10):4756-4762. doi: 10.3969/j.issn.1000-6923.2021.10.032FU S S, WANG Y, WANG X L, et al. Generation of extracellular antibiotic resistance genes during municipal wastewater chlorination and UV disinfection[J]. China Environmental Science,2021,41(10):4756-4762. doi: 10.3969/j.issn.1000-6923.2021.10.032 [81] 李金梅, 李曦, 张舒婷.消毒工艺对水体中抗生素抗性基因的去除效果[J]. 净水技术,2018,37(2):10-16. doi: 10.15890/j.cnki.jsjs.2018.02.003LI J M, LI X, ZHANG S T. Effect of disinfection process on removal of antibiotic resistance genes (ARGs) in water body[J]. Water Purification Technology,2018,37(2):10-16. doi: 10.15890/j.cnki.jsjs.2018.02.003 [82] ZHUANG Y, REN H Q, GENG J J, et al. Inactivation of antibiotic resistance genes in municipal wastewater by chlorination, ultraviolet, and ozonation disinfection[J]. Environmental Science and Pollution Research,2015,22(9):7037-7044. doi: 10.1007/s11356-014-3919-z [83] SHI P, JIA S Y, ZHANG X X, et al. Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water[J]. Water Research,2013,47(1):111-120. doi: 10.1016/j.watres.2012.09.046 [84] ZHANG T Y, HU Y R, JIANG L, et al. Removal of antibiotic resistance genes and control of horizontal transfer risk by UV, chlorination and UV/chlorination treatments of drinking water[J]. Chemical Engineering Journal,2019,358:589-597. doi: 10.1016/j.cej.2018.09.218 [85] 姚鹏城. 典型抗生素的预氧化降解: 可生化性及抑菌效应变化机制[D]. 上海: 上海师范大学, 2022. [86] MENG X Q, LI F J, YI L, et al. Free radicals removing extracellular polymeric substances to enhance the degradation of intracellular antibiotic resistance genes in multi-resistant Pseudomonas Putida by UV/H2O2 and UV/peroxydisulfate disinfection processes[J]. Journal of Hazardous Materials,2022,430:128502. doi: 10.1016/j.jhazmat.2022.128502 [87] HOU J, CHEN Z Y, GAO J, et al. Simultaneous removal of antibiotics and antibiotic resistance genes from pharmaceutical wastewater using the combinations of up-flow anaerobic sludge bed, anoxic-oxic tank, and advanced oxidation technologies[J]. Water Research,2019,159:511-520. doi: 10.1016/j.watres.2019.05.034 [88] RODRÍGUEZ-CHUECA J, della Giustina S V, ROCHA J, et al. Assessment of full-scale tertiary wastewater treatment by UV-C based-AOPs: removal or persistence of antibiotics and antibiotic resistance genes[J]. Science of the Total Environment,2019,652:1051-1061. doi: 10.1016/j.scitotenv.2018.10.223 [89] AHMED Y, ZHONG J X, YUAN Z G, et al. Simultaneous removal of antibiotic resistant bacteria, antibiotic resistance genes, and micropollutants by a modified photo-Fenton process[J]. Water Research,2021,197:117075. ◇ doi: 10.1016/j.watres.2021.117075 -




 下载:
下载: